Here one can find some visual excepts from my work (click on image to see the movie!). Some of these work were used in publications, some are still in progress and some never reached publisher (or I contribute only enough for credits section).
2013 Efficient simulation of multicellular systems
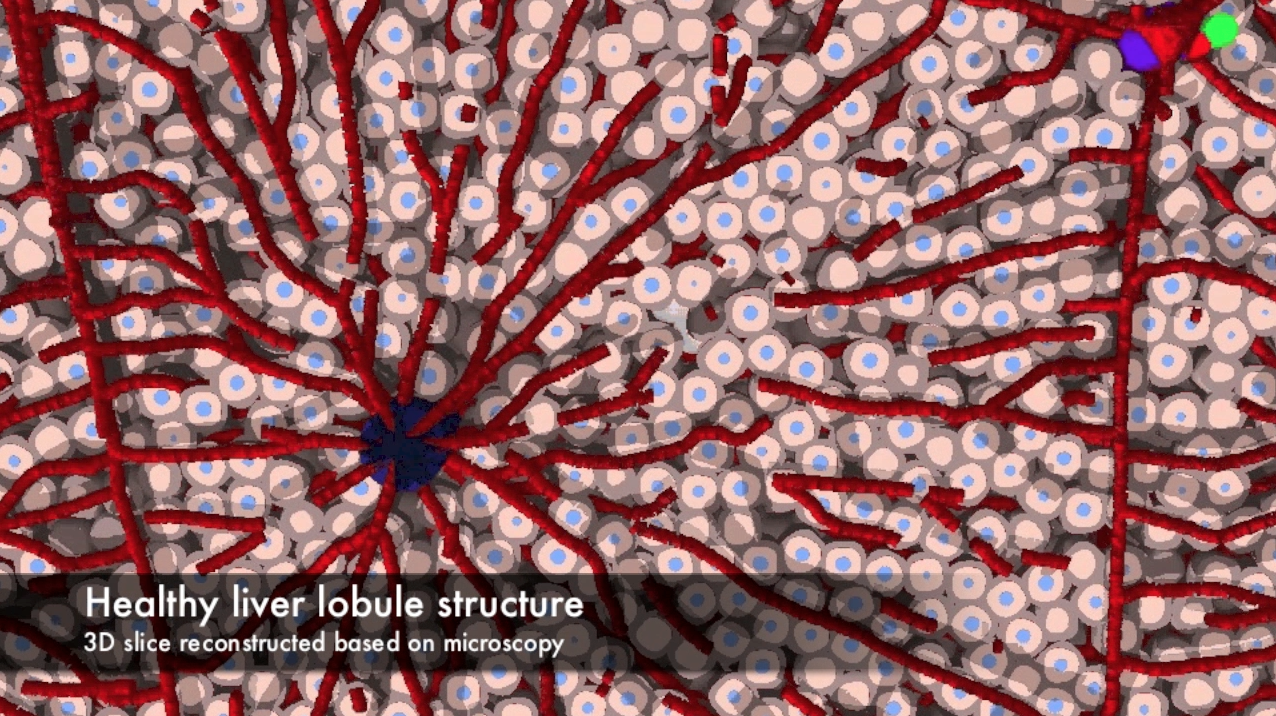
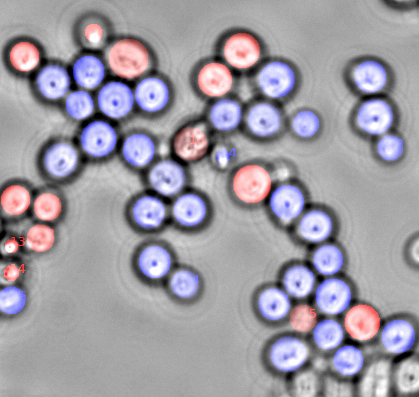
Agent-based, physical simulation of tumor growth inside of the liver. The framework for the simulations was created at INRIA Bang mostly by William Weens and extended by Francois Berataux under the supervision of Dirk Drasdo. The aim of the project is to perform cell based, mutiscale, biophysical simulations of tumor growth in the liver based on high dimensional microscopy imaging data. People: Francois Bertaux, William Weens, Dirk Drasdo, Stefan Hoehme, Szymon Stoma. Organizations: INRIA Bang (Paris, France), University of Leipzig / IZBI. Status: Unpublished.
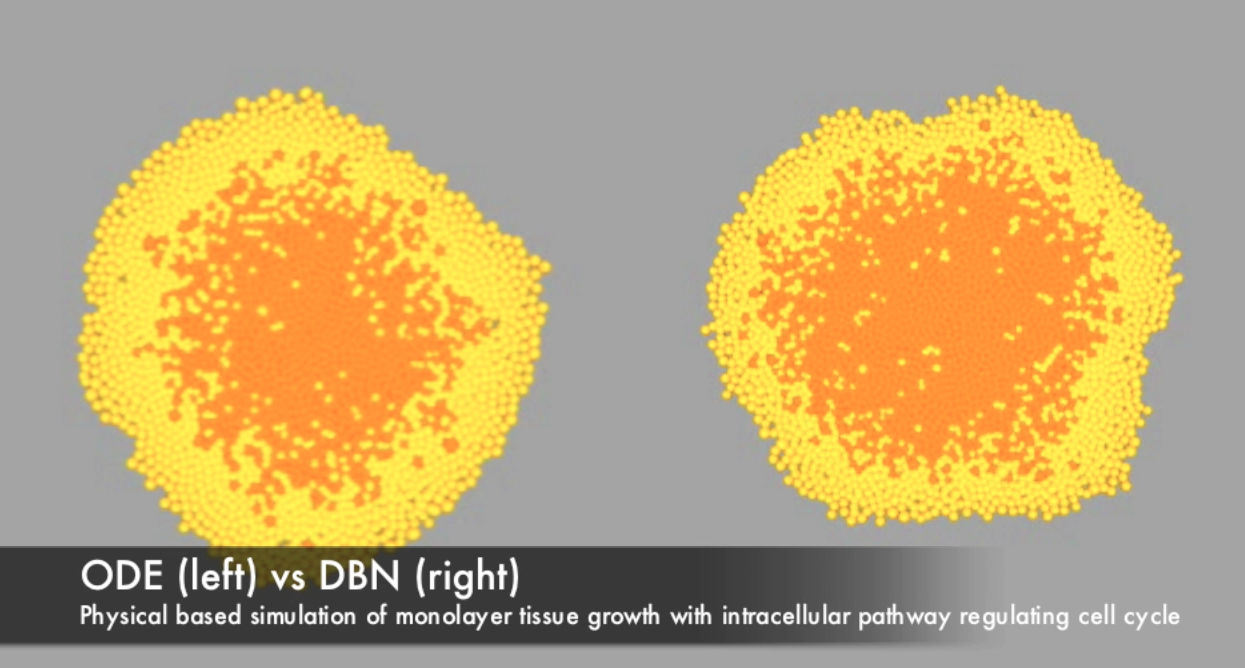
Agent-based, physical simulation of monolayer colony growth. The framework we created at INRIA Contraintes is based on the Dynamic Bayesian Networks used recently to estimate parameters for signaling pathways ("A Computational and Experimental Study of the Regulatory Mechanisms of the Complement System" by Liu Bing et al.). We decided to give DBN a try in the area of multicellular system simulations and indeed a significant speed up might be achieved without loosing too much precision (DBN is an approximation of ODE kinetics). People: Francois Bertaux, Gregory Batt, Dirk Drasdo, Hugues Asofa Zeufack, Liu Bing, P.S. Thiagarajan, Szymon Stoma. Organizations: INRIA Contraintes/Bang (Paris, France), School of Computing@National University of Singapore (Singapore). Status: Unpublished.
2012 Yeast growth image protocol,analysis and simulation algorithms
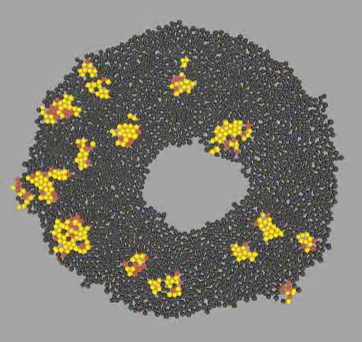
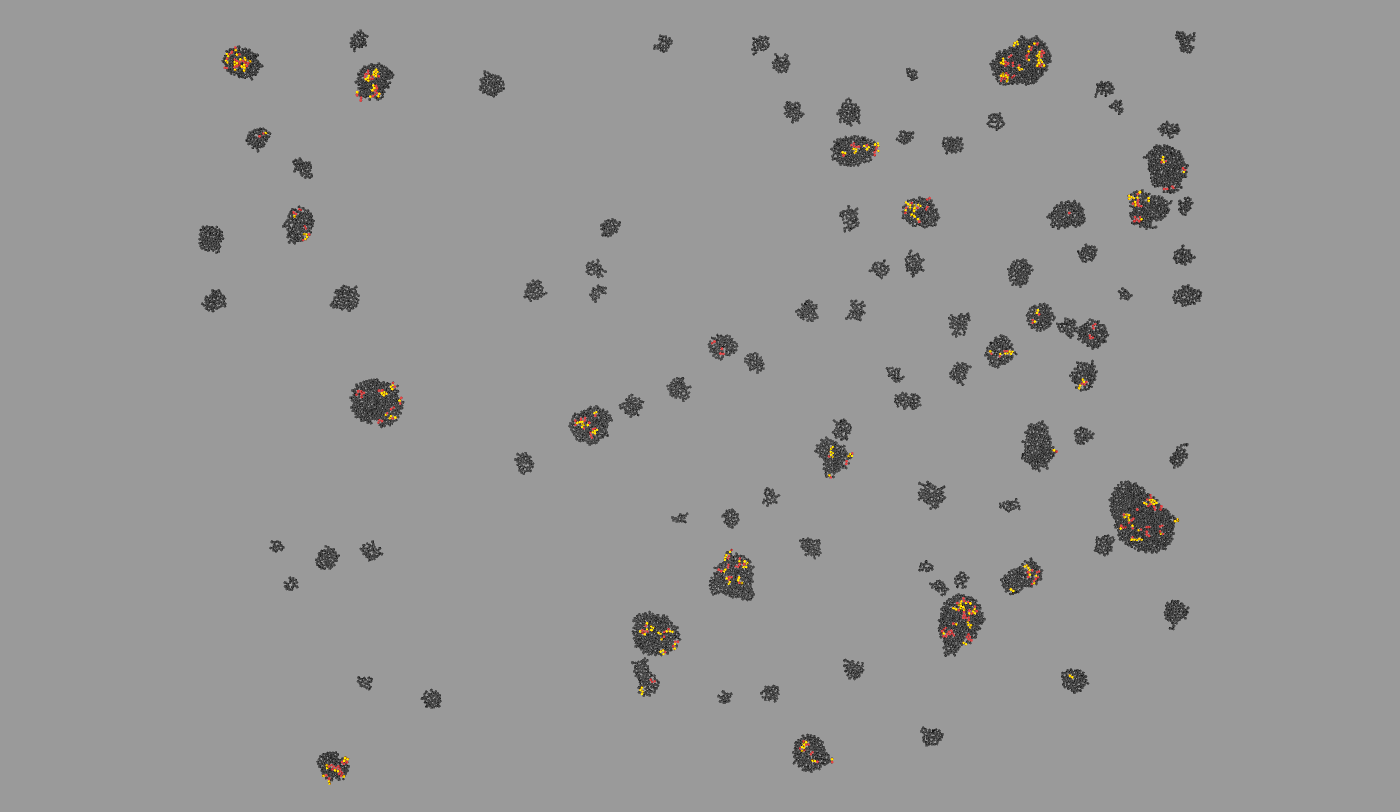
Agent-based, physical simulation of Yeast colony growth. Simulation in the context of Game of Life Synthetic Biology project realized together with MIT Ron Weiss lab and INRIA Bang Team. People: Francois Bertaux, Gregory Batt, Dirk Drasdo, Allen Lin, Jingjing Sun, Ron Weiss, Szymon Stoma. Organizations: INRIA Contraintes/Bang (Paris, France), Weiss Lab@MIT (MA, USA). Status: Unpublished.
Agent-based, physical simulation of Yeast colony growth. Simulation in the context of Game of Life Synthetic Biology project realized together with MIT Ron Weiss lab and INRIA Bang Team. People: Francois Bertaux, Gregory Batt, Dirk Drasdo, Allen Lin, Jingjing Sun, Ron Weiss, Szymon Stoma. Organizations: INRIA Contraintes/Bang (Paris, France), Weiss Lab@MIT (MA, USA). Status: Unpublished.
Yeast colony tracking algorithm developed in INRIA. People: Szymon Stoma. Organization: INRIA Contraintes (Paris, France) Status: Unpublished.
Single cell tracking from brightfield images algorithm developed in INRIA. People: Szymon Stoma, Filip Mroz, Adam Kaczmarek. Organizations: INRIA Contraintes (France), University of Wroclaw (Poland). Status: Unpublished.
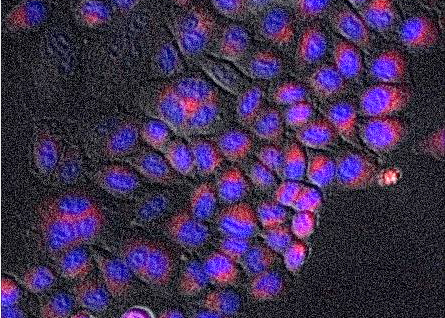
2012 HeLa apoptosis imaging protocol&analysis
Image acquisition and processing protocol developed in INRIA for the syne2arti project People: Xavier Duported, Francois Bertaux, Gregory Batt, Dirk Drasdo, Szymon Stoma. Organizations: INRIA Contraintes (France), Hersen Lab@University of Paris 5 (France). Status: Unpublished.
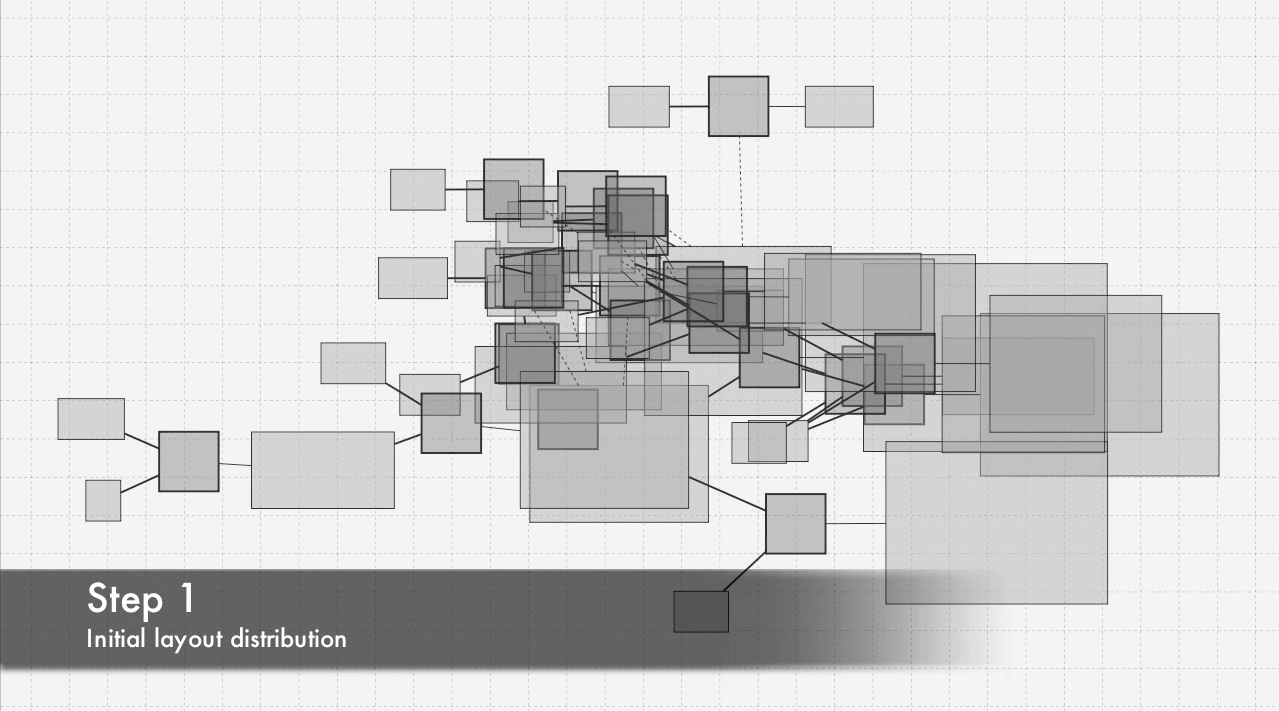
2011 Signaling pathway layout algorithm
Layout algorithm realized during Google summer school of code 2011. I was coauthor of the grant application. People: Thomas Handorf, Falko Krauze, Marvin Schulz, Max Flottman, Edda Klipp, Szymon Stoma. Organization: Klipp Lab@Humboldt University (Berlin, Germany). Status: Unpublished.
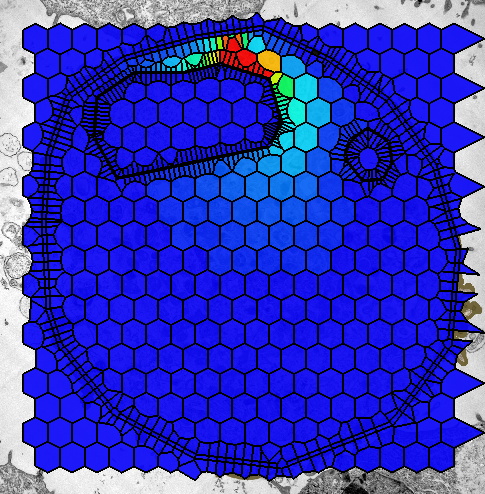
2011 STSE Spatio Temporal Simulation Environment
Spatio-temporal simulation environment (STSE): the overall goal of this project is to provide a software platform: a set of tools and workflows facilitating spatio-temporal simulations (preferably of biological systems) based on microscopy data. The framework currently contains modules to digitize, represent, analyze, and model spatial distributions of molecules in static and dynamic structures (e.g. growing). A strong accent is put for the experimental verification of biological models by actual, spatio-temporal data acquired using microscopy techniques. Organization: Klipp Lab@Humboldt University (Berlin, Germany). People: Martina Froehlich, Sussane Gerber, Edda Klipp, Szymon Stoma. Status: Published ("STSE: Spatio-Temporal Simulation Environment Dedicated to Biology", Stoma S, Froehlich M, Gerber S, Klipp E, BMC Bioinformatics, BMC Bioinformatics, 2011, 12(1):126.).
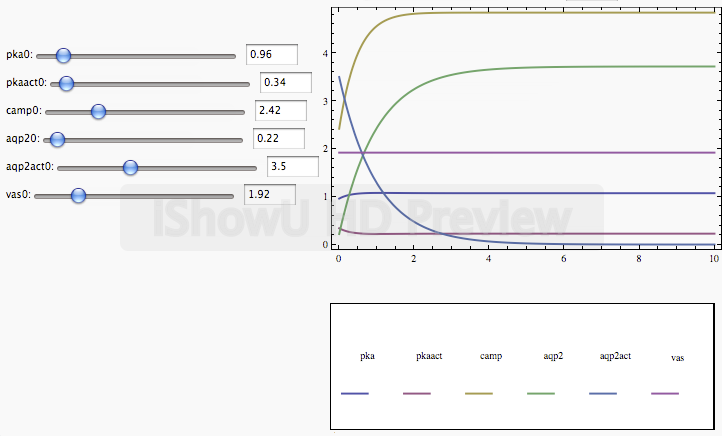
2010 Interactive ODE solvers
Interactive ODE solver developed to demonstrate systems behaviour developed for Aqua(glycero)porin project. people: Martina Froehlih, Edda Klipp, Szymon Stoma. Organization: Klipp Lab@Humboldt University (Berlin, Germany). Status: published ("A systems biology approach: modelling of aquaporin-2 trafficking", Fröhlich M, Deen PM, Klipp E., Genome Informatics).
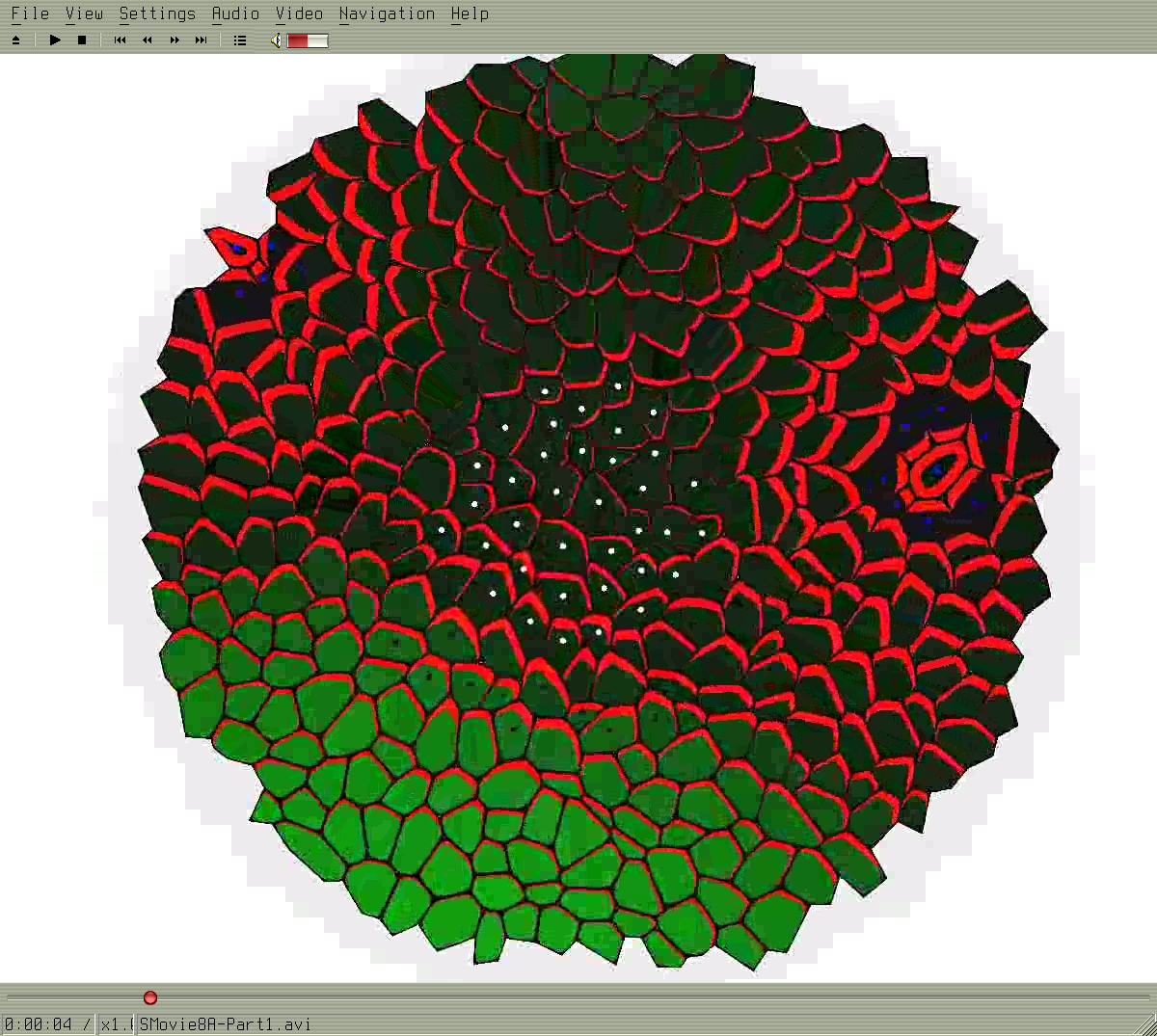
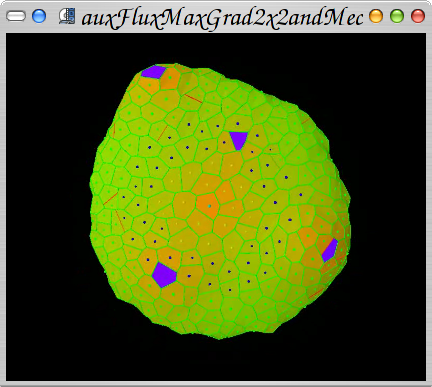
2008-2009 Growing meristem tissue with intercellular interactions
Formation of spiral phyllotaxis pattern. The inhibitory field concept is realized using the auxin concentration gradient. The auxin is distributed using an active transport. The active transport molecules (PINs) are distributed with concordance to the canalization hypothesis. People: Jerome Chopard, Christophe Godin, Jan Traas, Szymon Stoma. Organization: INRIA Virtualplants (Montpellier, France), Traas Lab@ENS (Lyon, France). Status: published ("Flux-Based Transport Enhancement as a Plausible Unifying Mechanism for Auxin Transport in Meristem Development",
Stoma S, Lucas M, Chopard J, Schaedel M, Traas J, Godin C, PLoS Computational Biology 4(10), 2008: e1000207 doi:10.1371/journal.pcbi.1000207 ).
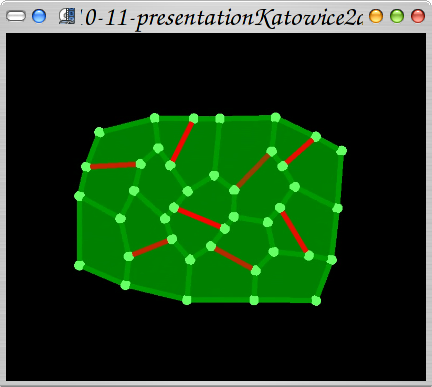
2006-2007 Meristem tissue physical based simulations
Simulation of growth of vegetal tissue based on mechanics: cells grow because of turgor, walls are viscoelastic and division occur when stress in a cell exceeds stress threshold. People: Jerome Chopard, Christophe Godin, Jan Traas, Szymon Stoma. Organization: INRIA Virtualplants (Montpellier, France), Traas Lab@ENS (Lyon, France). Status: published ("Using mechanics in the modelling of meristem morphogenesis",
Stoma S, Chopard J, Godin C, Traas J: Functional-structural plant models (FSPM), Napier, New Zealand, November 4-9, 2007.).
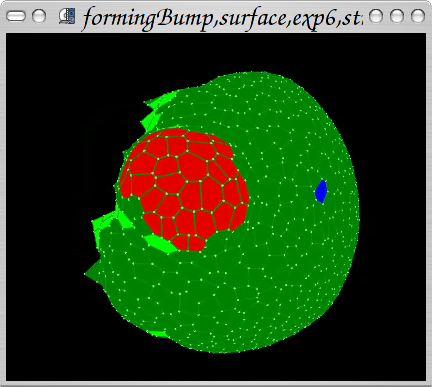
Coupling mechanics with physiology: reproduction of Arabidopsis thaliana PIN1 mutant behaviour (a plant with PIN1 knockout; it lacks the ability to actively transport growth hormon auxin required for organ initiation). A hormon is therefore applied externally (in experiment: with lanoline paste). After application the growth rate is increased locally leading to bulging effect. Around the "bump" a ring of cells with increased stress can be identified. Auxin: red. Light green: hypothetical stress gene expressed. People: Jerome Chopard, Christophe Godin, Jan Traas, Szymon Stoma. Organization: INRIA Virtualplants (Montpellier, France), Traas Lab@ENS (Lyon, France). Status: published ("Using mechanics in the modelling of meristem morphogenesis",
Stoma S, Chopard J, Godin C, Traas J: Functional-structural plant models (FSPM), Napier, New Zealand, November 4-9, 2007.).
First steps in coupling mechanics with phyllotactic patterns. The idea is to express feedback loop of morphogen transport with the growth. People: Jerome Chopard, Christophe Godin, Jan Traas, Szymon Stoma. Organization: INRIA Virtualplants (Montpellier, France), Traas Lab@ENS (Lyon, France). Status: published ("Using mechanics in the modelling of meristem morphogenesis",
Stoma S, Chopard J, Godin C, Traas J: Functional-structural plant models (FSPM), Napier, New Zealand, November 4-9, 2007.).